ECU biology professor researches genetic markers associated with sudden infant death syndrome
The loss of a child is an emotionally sensitive experience for a parent, no matter the age, but when the cause is unknown, as in the case of sudden infant death syndrome (SIDS), there may be more questions than answers.
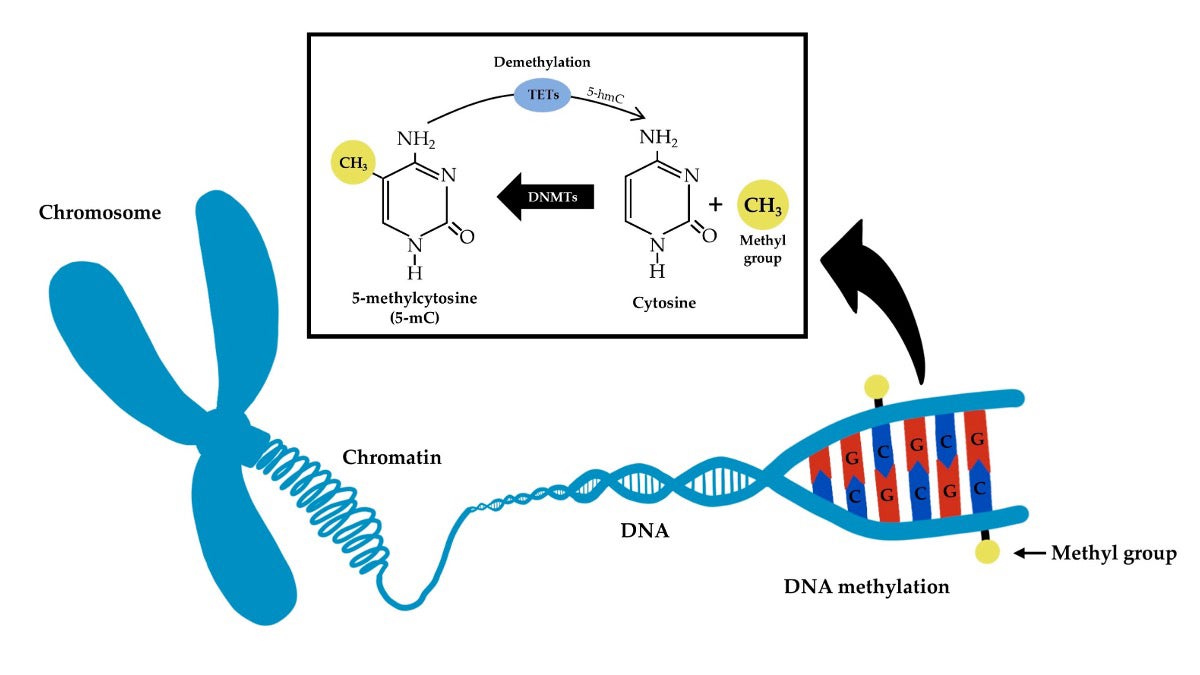
Special proteins called DNA methyltransferases add small chemical tags called methyl groups (CH3) to certain parts of DNA (specifically to cytosine) in a process called DNA methylation. They can be removed through a process called demethylation. This process can affect how genes are turned on or off and what they do. (Contributed diagram)
However, over the past 15 years, East Carolina University’s Dr. Keith Keene, Thomas Harriot College of Arts and Sciences associate dean for research and professor in the Department of Biology, has been trying to identify what types of biomarkers in genes, blood and tissues may lead to an increased risk of SIDS.
After conducting his postdoctoral research at the University of Virginia School of Medicine, Keene and other colleagues were approached by a family who had lost a child to SIDS and wanted to fundraise and contribute to SIDS research.
“SIDS is the second leading cause of death — closely behind congenital and genetic abnormalities — in infants from 1 month to 1 year of age,” Keene said. “SIDS is the sudden unexpected death of an infant that remains unexplained after autopsy, death scene investigation and review of medical history.”
“Our research aims to identify biomarkers for SIDS diagnosis and risk, where we can not only identify and prevent SIDS deaths but also use genomics as a diagnostic approach to perform molecular autopsies for SIDS,” he said.
According to the Centers for Disease Control and Prevention approximately 1,500 infants in the United States died of SIDS in 2022 despite a significant decline in incidents since the Back to Sleep risk reduction campaign. The campaign, named for the recommendation to place babies on their backs to sleep (now known as the Safe to Sleep campaign), was launched by the National Institutes of Health in 1994.
In the spring of 2020, along with Fern Hauck and Josyf Mychalecky, two colleagues at the University of Virginia, Keene and his team were awarded a four-year, $2.3 million National Institutes of Health/National Institute of Child Health and Human Development grant to continue research into the cause of SIDS. The grant project, which moves the research field away from environmental factors into the potential biological risk factors, was extended through summer 2025.
Metabolomics is the scientific study of the chemical processes involving metabolites, the small molecules present within an organism, cell or tissue. This image shows the application of metabolomics across science and medicine. (Contributed by Metabolon Inc.)
“Extensive research has identified several epidemiological risk factors for SIDS, and more recent research has started to provide evidence of a potential role for genetic susceptibility in the pathophysiology of the disorder,” Keene said.
The goal of their study — one of only a handful and currently the largest research study of its kind — is to meticulously evaluate genetic expression, and identify DNA methylation (which prevents genes from being switched on) and metabolomics (metabolites) profiles that might serve as biomarkers in infants at greater risk of death from SIDS.
“I think this underscores the complex biological nature of SIDS. Many key biological pathways and processes are important, or have potential implications, for SIDS,” Keene said. “It is also important to understand that there are many potential sub-types of SIDS. It’s not a one-size-fits-all diagnosis. Some infants may die of SIDS because of brain or cardiac issues; others it may be respiratory issues or it could be a combination of all three.”
Through their study, the researchers examined liver, heart and blood samples from approximately 300 cases of SIDS. They used an omics approach, which encompasses next-generation sequencing (exome plus RNA), genome-wide epigenetics (DNA modification), and profiling of metabolomics (small molecules or metabolites produced by cells) to identify contributors for SIDS.
Earlier this year, the team published its first paper, announced through a University of Virginia School of Medicine press release. The paper, “Metabolomic profiles of infants classified as sudden infant death syndrome: A case-control analysis,” which appears in the scientific journal eBioMedicine, identified 35 biomarkers in an infant’s blood that may be important biomarkers for SIDS.
More research is needed, though, and Keene will continue to work with his former colleagues through the summer to analyze more omics data for associations with SIDS.
Keene said these findings could lead to diagnostic tests, prescreening and early detection of high-risk infants, resulting in preventative measures to eliminate SIDS.
“I think there is nothing worse than losing a child,” he said. “So if we can find ways to either identify high-risk infants or prevent SIDS altogether, that will not only save lives but contribute to the reduction of the emotional impact that losing a child can have on a family.”
